The Secret Between DNA Methylation and Cancer
DNA methylation is one of the most intensively studied epigenetic modifications in mammals. In normal cells, DNA methylation can effectively regulate gene expression levels, and the inactivation of some tumor suppressor genes is caused by hypermethylation of promoter regions. A large number of studies have shown that DNA methylation can lead to extensive gene silencing in most cancer species. In addition to alterations in methylation levels in promoter regions and DNA repeats, methylation is also associated with regulation of expression of non-coding Rnas, such as micrornas associated with tumor inhibition. The link between DNA methylation levels and tumorigenesis encourages us to decode the human epigenome. This article will take you to understand the relationship between DNA methylation and cancer.
What is methylation?
In mammalian cellsDNAThe methylation is inDNAmethyltransferase(DNMT)Under the catalysis of cytosine bases(5 -Methyl cytosine;5mC)Carbon of- 5The addition of methyl groups to the site usually occurs in cytosine-Guanine dinucleotide(CpG)In. In the human genome, there are about2800Ten thousandCpGLoci, and theseCpGLoci are not evenly distributed, and in some regions of the genome,CpGAt or above normal probability.CpGIslands are mainly located in the promoter and first exon region of genes, and their function is to regulate the expression of downstream genes through methylation and demethylation. ifCpGHypermethylation occurs in islands, and gene expression is inhibited. mostCpGThe length of the island is500-1000.Student: Base pair(bp)Unlike most of the genome is located inCpGintra-islandCpGSites are usually unmethylated in normal somatic cells.
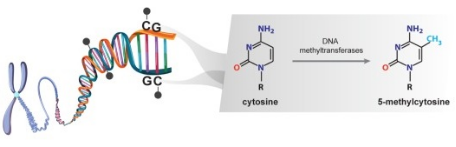
Figure 1 Principle of DNA methylation (Skvortsova K et al.,2019[1])
DNA methyltransferases involved in this process include DNMT1, DNMT3A and DNMT3B. DNMT1 recognizes semi-methylated DNA and maintains existing methylation patterns. DNMT3A and DNMT3B add methyl groups to unmethylated cytosine. In addition to 5-methylcytosine, 5-hydroxymethylcytosine (5hmc), as a methylated intermediate of cytosine, has been identified as the two most common epigenetic markers. The 5mC is oxidized to 5hmc by TET proteins of the dioxygenase family [2], and the 5mC and 5hmc levels of DNA play an important role in the occurrence and development of tumors.
Methylation and cancer
DNAMethylation is essential for maintaining the normal function and development of cells. The characteristics of methylation in tumor cells are very different from those in normal cells. Changes in both low and high methylation levels can be detected in cancer cells. In fact, during the development of the tumor, the apparent level is already abnormal,DNAThe methylation changes as a whole. In general,CpGAn overall decrease in methylation levels in the region can lead to genomic instability, but less activation of silent proto-oncogenes.
It is well known that the development and progression of cancer is accompanied by changes in DNA methylation patterns, including DNA hypomethylation of reverse transcriptional elements, centromeres, and proto-oncogenes, as well as methylation of key gene regulatory elements associated with gene suppression, such as overlapping regions of distal enhancer and promoter transcription initiation. As shown in Figure 2, patterns of DNA methylation differ widely between normal and cancer cells across all gene regulatory elements of the genome. Most CpG sites in the normal genome carry 5mC, and the distal enhancer element and CpG island region are resistant to DNMT activity. Cancer cells are characterized by overall loss of methylation genetic modification and abnormal methylation sites in enhancer and promoter regions. This change in the distribution of methylation leads to the inhibition of the expression of tumor suppressor genes, along with the increase of proto-oncogene expression, which further promotes the occurrence and development of tumors.
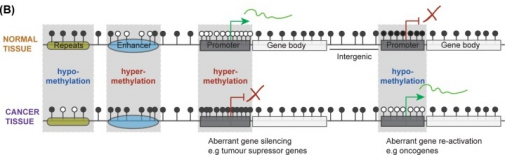
Figure 2 Differences in DNA methylation patterns between normal and cancer cells (Skvortsova K et al.,2019[1])
DNA methylation will become a common tumor marker
Given that normal cells and cancer cellsDNAThere are significant differences in methylation patterns that makeDNAMethylation has the potential to be a screening indicator. Therefore,DNAMethylated biomarkers must overcome current limitations and be more sensitive and specific than existing therapies or be more acceptable by minimally invasive methods. Numerous studies have shown that in cancer diagnosisDNAMethylation has been significantly improved in predicting the prognosis of patients, guiding the selection of treatment regiments, and monitoring the therapeutic effect.
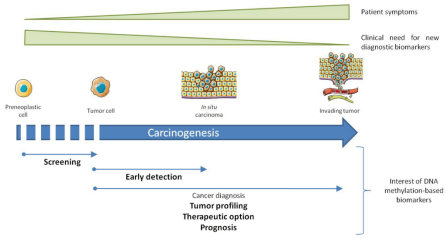
Figure 3 Significance of DNA methylation biomarkers in cancer diagnosis (Delpu, Y et al.,2013 [3])
1. Early diagnosis of cancer
Early diagnosis of cancer is very important for the treatment and prognosis of patients. McCluskey et al. showed that p16 gene hypermethylation difference could distinguish benign and malignant ovarian tumors, and the distal promoter was methylated in 33% of low-grade potential malignant tumors, compared with 5% in cancer [4]. Similarly, another study has shown that the methylation level of the gene encoding miR-148a is a potential diagnostic tool for differentiating pancreatic cancer from chronic pancreatitis [5].
2. Cancer classification
DNA methylation can be used for cancer typing. There are differences in DNA methylation patterns between different types of cancer, so DNA methylation patterns can be a powerful tool for improving tumor classification. For example, biomarkers based on high-resolution DNA methylation (67,487 probes) were used in 49 patients with acute lymphoblastic leukemia (ALL) to distinguish different subtypes of ALL and influence clinical outcomes [6].
3. Cancer prognosis assessment
In addition to early diagnosis and cancer typing, DNA methylation can predict tumor response to therapy and improve patient outcomes. For example, DNA hypermethylation of the MGMT promoter in 40% of glioma patients is directly related to tumor resistance to conventional chemotherapy based on alkylating agents. In addition, the accumulation of normal cells in the tumor can affect the evaluation of MGMT expression in the tumor. More importantly, it is associated with tumor regression and extended overall survival and disease-free survival [7].
4. Cancer treatment monitoring
DNA Methylation can be used to monitor the effects of cancer treatments, such as chemotherapy, radiation, etc., according toDNAChanges in methylation levels to assess therapeutic effect. For patients with lymphoma, after treatmentDNAChanges in methylation levels can be used to assess the effect of treatment.
Advantages of DNA methylation
1, high tissue specificity: Epigenetic changes usually occur early in the tumor and are tissue and cancer type specific. Different tissue and cell typesDNAMethylation patterns differ and can be used to determine the type and origin of the tumor. Therefore, the methylation status of related genes is an early sensitive indicator of tumorigenesis and can be considered as a promising tumor marker.
2. High stability: Compared with other biomarkers, such as proteins and RNA, DNA methylation is more stable during sample collection, transportation and storage, which can better maintain its integrity and reliability.
3. High sensitivity: DNA methylation tests can detect abnormal DNA methylation patterns at very early stages of tumor development, even before tumor formation.
4. High accuracy: DNA methylation test has high accuracy and repeatability, and can be used for early diagnosis, typing, grading and prognosis of tumors.
5. High diversity: DNA methylation detection can be applied to various sample types, including blood, urine, tissue sections and body fluids, etc., which has a wide range of application prospects.
Several methods for detecting methylation of specific DNA fragments
1. Methylation-specific PCR (MS-PCR)
This method is economical, without special instruments, and is now the most widely used. The main process is methyl conversion followed by primer specificPCR.MS-PCRMedium need design2For primers, detectMSPThe amplified product is methylated or unmethylated according to the treatmentDNAWhether the primers of the chain can amplify fragments to determine whether the tested site exists methylation.
2. Fluorescence quantification (Methylight)
The principle is to use a fluorescent hydrolysis probe inMSPThe amplification also detected fluorescence intensity, making it possible to quantitatively detect methylation. It's treated with bisulfiteDNAFragment, and design a complementary probe with the site to be measured to carry out real-time quantificationPCR. Its advantages are high throughput, high sensitivity, and rapid analysis of multiple locus and multiple gene loci. But the law is expensive.
3. Methylation-sensitive restriction endonucase PCR (MS-RE)/Southern method
In this method, DNA was digested into fragments of different sizes using the non-cleavage of methylated regions by methylation-sensitive restriction endonucliase. The cost of this method is low, the methylation site is clear, and the results are easy to interpret. However, this method has a large demand for samples and is not suitable for mixed samples. Incomplete digestion of enzymes may cause false positivity.
4. Gradient Gel electrophoresis of methylation Sensitivity Denaturation (MS-DGGE)
The principle isDNAThe denaturant concentration in the gel increases from top to bottom when the fragment is electrophoresis with denaturant gradient polyacrylamide gelDNAThe segment reaches the unchained regionTAt a certain concentration of the same value,DNAThe unchained chain becomes branched, moves slower, and stays in one place in the gel, but it's differentDNASegments are separated. This method does not need to be known in advanceCpGSite and sample sequence, the sample quantity is small, but the unchained temperature andDGGEThe denaturation concentration gradient needs to be fumbled and is easy to be missed.
Early tumor screening is an effective means to reduce the mortality of malignant tumors and is the development direction of detection technology in the future. DNA methylation detection has the advantages of improving the early tumor detection rate, simple sampling, convenient non-invasive, high sensitivity and high specificity, which can effectively reduce the incidence and mortality of malignant tumor in our country. Thus, the application prospect of DNA methylation in the field of tumor will be very broad!
References:
1. Skvortsova K, et al. The DNA methylation landscape in cancer. Essays Biochem. 2019 Dec 20; 63 (6) : 797-811.
2.Giorgia Gurioli. Epigenetic Characterization of Cell-Free DNA. Cell-free DNA as Diagnostic Markers pp 129-135.
3.Delpu, Y.; Cordelier, P.; Cho, W.C.; Torrisani, J. DNA Methylation and Cancer Diagnosis. Int. J. Mol. Sci. 2013, 14, 15029-15058.
4.McCluskey, L.L.; Chen, C.; Delgadillo, E.; Felix, J.C.; Muderspach, L.I.; Dubeau, L. Differences inp16Gene methylation and expression in benign and malignant ovarian tumors. Gynecol. Oncol 1999, 72, 87-92.
5.Hanoun, N.; Delpu, Y.; Suriawinata, A.A.; Bournet, B.; Bureau, C.; Selves, J.; Tsongalis, G.J.; Dufresne, M.; Buscail, L.; Cordelier, P.; et al. The silencing of microRNA 148a production by DNA hypermethylation is an early event in pancreatic carcinogenesis. Clin. Chem 2010, 56, 1107-1118.
6.Stumpel, D.J.P.M.; Schneider, P.; van Roon, E.H.J.; Boer, J.M.; de Lorenzo, P.; Valsecchi, M.G.; de Menezes, R.X.; Pieters, R.; Stam, R.W. Specific promoter methylation identifies different subgroups of MLL-rearranged infant acute lymphoblastic leukemia, influences clinical outcome, and provides therapeutic options. Blood 2009, 114, 5490-5498.
7.Esteller, M.; Garcia-Foncillas, J.; Andion, E.; Goodman, S.N.; Hidalgo, O.F.; Vanaclocha, V.; Baylin, S.B.; Herman, J.G. Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. N. Engl. J. Med 2000, 343, 1350-1354.

