Molecular diagnostics ----IVD industry proud
![]()
Technical classification
...
...
...
At present, molecular diagnostic technology mainly includes nucleic acid molecular hybridization technology, nucleic acid amplification technology, gene chip technology, gene sequencing technology, has been widely used in clinical departments.
Molecular diagnostic technology based on nucleic acid molecular hybridization
Nucleic acid molecular hybridization technology refers to the specific formation of two single strands of nucleic acid from different sources by the principle of base complementary pairing under certain conditions. They include Southern blot hybridization, Northern blot hybridization, in-situ hybridization and fluorescence in situ hybridization (FISH). FISH technology is used to form hybridization between fluorescent-labeled probe and target fragment, and obtain relevant data of DNA to be tested through fluorescence detection system. It has the characteristics of strong specificity and fast detection speed, and is widely used in prenatal diagnosis [1]. In 1993, the American Society of Medical Genetics published relevant instructions on the application of FISH in prenatal diagnosis [2], and this technique has become an expert consensus as a supplement to karyotype analysis in the field of prenatal diagnosis.
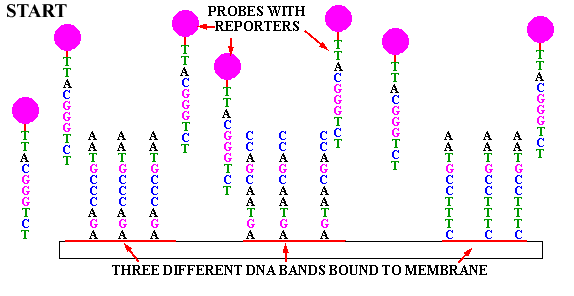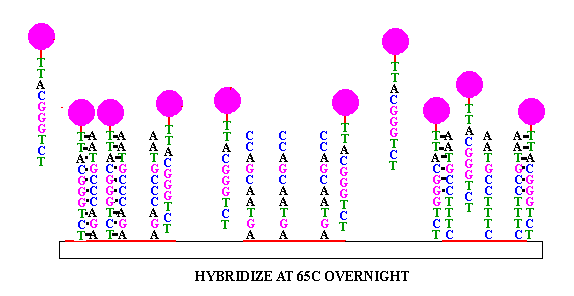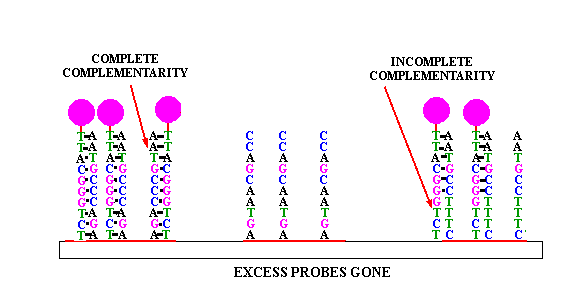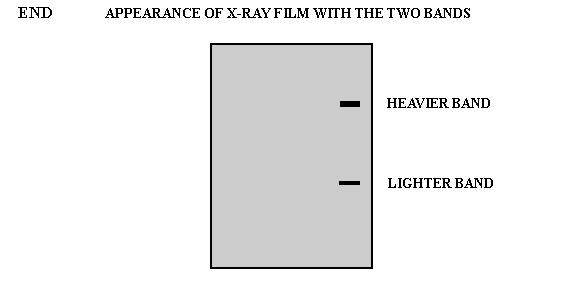
Figure 1 Principle of nucleic acid molecular hybridization technology (Source: Network)
Molecular diagnostic techniques based on nucleic acid amplification
In the 1980s, Mullis developed polymerase chain reaction (PCR), a technique for in vitro amplification of target nucleic acid fragments. Nucleic acid amplification technology is widely used in clinic, and a variety of PCR technologies have been developed so far, including quantitative real-time PCR(qPCR) and digital PCR(dPCR). Because the operation process of qPCR is carried out in a closed system, the contamination probability is reduced, and quantitative detection can be carried out by monitoring fluorescence signals. Therefore, the clinical application of QPCR is the most widespread, and it has become the leading technology in PCR [3]. dPCR is also known as single-molecule PCR. Its principle is to place a single nucleic acid molecule in an independent reaction unit for PCR amplification, then detect the fluorescence at the end of the reaction chamber and carry out molecular counting statistics to achieve quantitative analysis. Due to the characteristics of high sensitivity and accuracy, it is not easy to be interfered by PCR reaction inhibitors, and absolute quantification in real sense can be achieved without standard substances, which has become a research hotspot [4].

Figure 2. Principle of PCR technology (Source: Andy Vierstraete,1999)
Molecular diagnostic technology based on gene chip technology
Gene chip technology is to fix a large number of nucleic acid probes with known sequences on the substrate surface, then hybridize them with target nucleotides, and obtain the sequence information of the sample to be tested through the detection of the probes. According to the types of probes, gene chips can be divided into two categories, namely cDNA microarray chips and oligonucleotide microarray chips [5]. The probe of the former is the corresponding cDNA of the specific mRNA in the cell, while the probe of the latter is the oligonucleotide fragment synthesized in situ. Gene chip technology is widely used in genotyping, gene expression and single nucleotide polymorphism detection due to its high automation, simple operation and high throughput.
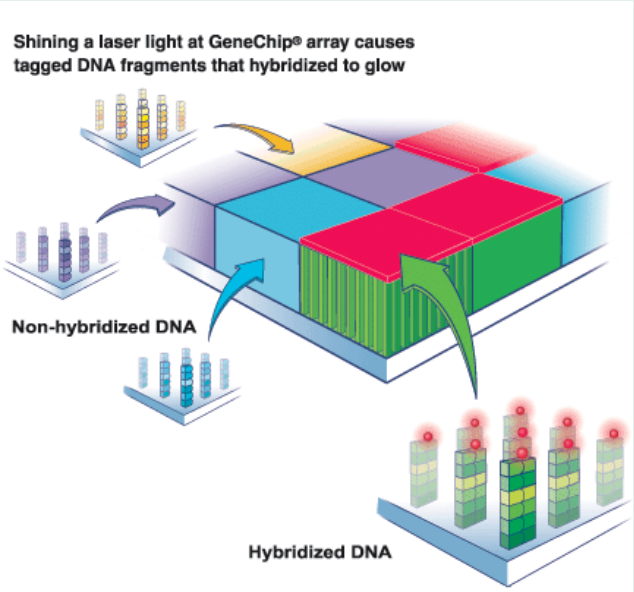
Figure 3 Principle of gene chip detection (Source: Network)
Molecular diagnostic technology based on gene sequencing technology
Gene sequencing technology started from the DNA dideoxy end termination sequencing method invented by Sanger in 1977. Because of its high accuracy, it is considered as the "gold standard" of gene sequencing. However, its clinical application is limited by its high cost and low throughput.
Since entering the 21st century, gene sequencing technology has developed rapidly, and the second and third generation sequencing technology has been invented successively. The second generation sequencing technology is also known as high throughput sequencing (HTS) technology. Compared with the first generation sequencing, it can achieve large-scale parallel sequencing. The basic principle is to segment the genome into short fragments, sequence short fragments and then splicing. It has many advantages such as short detection time, high sensitivity, high throughput and economy, which greatly promotes the application of molecular diagnostics in clinical detection and advances the development of genomics. HTS technology is the most widely used sequencing technology in clinic. The third-generation sequencing technologies are mainly represented by single-molecule sequencing and Nanopore sequencing, such as Min ION nanopore sequencing technology developed by Oxford Nanopore, SMRT sequencing technology developed by Pac Bio, etc. [6]. It does not require nucleic acid amplification, reading length is obviously better than HTS technology, and can assist personalized medicine at the genome level, which is the main technical direction of future development.

Figure 4 DNA sequencing using Illumina platform
(Source: J. M. Churko,et al., 2013[7])
Clinical application
...
...
...
With the continuous development of molecular diagnostic technology, it has become a new clinical means and ideas for effective disease diagnosis. Especially in the fields of infectious diseases, tumors and genetic diseases, it not only effectively makes up for the deficiency of traditional methods, but also provides important evidence for disease prevention, diagnosis, treatment monitoring and prognosis assessment.
Clinical application in tumor
The occurrence of tumor is the result of the joint action of genes and environment. The individualized and precision medicine of malignant tumor is of great significance. Molecular diagnostic technology not only contributes to the prevention and early diagnosis of cancer, but also plays a role in the molecular classification, prognosis prediction and treatment program screening of cancer.
FISH technology is used to detect changes in the number and structure of chromosomes, and the status of chromosomes is closely related to the development of tumors. The detection of specific chromosomes by FISH technology can predict the prognosis of cancer. Mian et al. [8] conducted long-term follow-up of 81 patients with uroepithelial carcinoma (UC), and used multicolor fluorescence in situ hybridization to detect 9p21(p16) sites and chromosomes 3, 7 and 17 of the patients, so as to screen those at high risk of bladder cancer recurrence and predict the development of the disease.
In nucleic acid amplification technology, fluorescence quantitative PCR has been widely used in tumor therapy for its unique advantages. Squamous cell carcinoma of the head and neck (HNSCC) is one of the most common cancers, and several studies have shown that human papillomavirus (HPV) infection is one of the pathogenic factors. Polanska et al. [9] collected tissue samples from 74 primary HNSCC patients and analyzed 15 HNSCC-related molecular markers by real-time quantitative polymerase chain reaction (qRT-PCR). Meanwhile, PCR was applied to detect HPV. Studies have confirmed differences in the expression of common biomarkers MT2A, MMP9, FLT1, VEGFA and POU5F in HPV-negative and HPV-positive HNSCC patients, which has important significance for predicting disease development and survival in HNSCC patients.
At the same time, HTS technology is also widely used in liquid tumor biopsies due to its high sensitivity. Lung cancer is the cancer with the highest incidence among Chinese men, among which the most common type is non-small cell lung cancer (NSCLC). In the targeted treatment of NSCLC, gene mutation detection is essential. Dono et al. collected blood samples from 42 patients and used HTS to detect mutations in exon T790M of EGFR-country 20 in plasma ct DNA to assess whether patients who had developed resistance to first - and second-generation EGFR-TKI therapy could continue to receive oxitinib, the third-generation EGFR-TKI drug. This study found that due to the extremely high sensitivity and detection rate of HTS, liquid biopsy could more effectively replace tissue detection and improve patient compliance [10].
Clinical application in genetic diseases
Statistics from the China Report on the Prevention of Birth Defects show that about 1 million babies are born with birth defects every year in China, and heredity is one of the factors causing birth defects. Genetic diseases are diseases caused by changes in genetic material or control of pathogenic genes, which are generally divided into chromosome syndrome, single gene inherited diseases and polygene inherited diseases. In recent years, molecular diagnostic technology has played an important role in genetic disease diagnosis, newborn screening and other fields.
As a new technology, gene chip is gradually replacing karyotype analysis in prenatal diagnosis. aCGH technique can rapidly detect chromosome CNV and the source of variation at the whole genome level. Chong et al. applied aCGH technology for prenatal diagnosis of more than 1000 fetuses with abnormal ultrasound examination. Due to the high resolution of aCGH chip technology, its combined application with ultrasound examination will improve the accuracy of detection [11].
High-throughput sequencing technology can sequence hundreds of thousands to millions of DNA molecules at a time, and is one of the most popular technologies at present. For the detection of chromosome aneuploidy disease, maternal cfDNA can be sequentially sequed by HTS, and the differences of different chromosomes in peripheral blood can be analyzed using biological information tools, so as to accurately identify the changes in fetal chromosomes. Bianchi et al. applied HTS to conduct large-scale parallel sequencing and detected trisomy 21 and 18 in blood samples of 1,914 pregnant women from 21 centers in the United States. Compared with traditional screening methods, the false positive rate of HTS technology decreased significantly and the accuracy rate was high [12].
Clinical application in infectious diseases
Infectious diseases are diseases caused by human infection with bacteria, viruses, fungi and other pathogenic microorganisms. According to the WHO, 17 million people die each year from infectious diseases. Traditional pathogen diagnosis is time-consuming and less accurate, while molecular diagnosis is faster and more accurate, enabling early treatment and virus control, which is the main development trend of pathogen detection.
PCR technology can not only detect virulence factors, pathogenic factors or variation factors in diseases, but also determine the DNA and RNA of pathogens. In addition to detecting the cause of diseases, PCR technology can also conduct genotyping and drug resistance detection of pathogens, so it is widely used. The pathogen detected by conventional PCR is single, so it is difficult to detect various mixed infections in clinic. Multiple PCR(mPCR) can solve this problem. Zhang Hailin et al. used mPCR technology for rapid detection of pathogens of lower respiratory tract infections in children, which is of high clinical value for guiding drug use in later period [13].
Molecular diagnosis has become an important subject to solve many scientific and medical problems. It has been widely used in clinical practice to promote disease prevention, diagnosis, prognosis prediction and the formulation of individualized medical programs. With the aging of the Chinese population, the incidence of cancer and chronic diseases will continue to rise, and the national health awareness is also gradually improving. Molecular diagnostics will be automated to reduce labor consumption and cost, and automated interpretation of test results will also reduce manual errors. At the same time, the breakthrough of single molecule technology also makes the detection of high sensitivity become the future development trend of molecular diagnostic technology, improve the detection efficiency. In addition, non-invasive detection and minimally invasive detection will improve patient compliance, which is conducive to the further promotion and application of molecular diagnostic technology in clinical practice. In the next few years, molecular diagnostic technology will be more widely used in physical examination, early diagnosis, prevention and treatment of major diseases.
Reference literature
[1] Liu Wantong, Tong Mei, Lin Fuyu et al. Advances in clinical application of molecular diagnostic techniques [J]. Biotechnology Communications,2020,31(02):240-250. (in Chinese)
[2] American College of Medical Genetics.Prenatal interphase fluorescence in situ hybridization(FISH)policy statement[J].Am J Hum Genet,1993,53(2):526.
[3] Yuan Yinchi, Zhao Xiaoqin, Chen Daming, et al. Research and development of in vitro diagnostic reagents and market development [J]. Biotechnology,2017,(4):16-24. (in Chinese)
[4] Lin Jiaqi, Su Guocheng, Su Wenjin, et al. Research progress of digital PCR technology and its application [J]. Chinese Journal of Bioengineering,2017,(2):170-177.
[5] Liu Tianlong, Li Yuwen, Zhang Yikai, et al. Application of gene chip technology in the study of pharmacological mechanism of traditional Chinese medicine [J]. Chinese Pharmacy,2016,27(28):4021-402. (in Chinese)
[6] Liu Yanhu, Wang Lu, Yu Li, et al. The principle and application of single molecule real-time sequencing [J] Genetics, 2015,37(3):259-268. (in Chinese)
[7] J. M. Churko, G. L. Mantalas, M. P. Snyder and J. C. Wu, Overview of High Throughput Sequencing Technologies to Elucidate Molecular Pathways in Cardiovascular Diseases, Circulation Research, 2013:112(12), 1613-1623.
[8] Mian C, Comploj E, Resnyaket E,et al. Long-term follow-up of intermediate-risk non-muscle invasive bladder cancer sub-classified by multi-coloured FISH[J].Anticancer Res,2014,34(6):3067-3071.
[9] Polanska H, Heger Z, Gumulec J, et al. Effect of HPV on tumor expression levels of the most commonly used markers in HNSCC[J].Tumor Biol, 2016, 5 (6) : 7193-7201.
[10] Dono M, De Luca G, Lastraioli S, et al. Tag-based next generation sequencing:a feasible and reliable assay for EGFR T790M mutation detection in circulating tumor DNA of non small cell lung cancer patients[J].Mol Med, 2019,25(1):15.
[11] Chong H P, Hamilton S, Mone F,et al. Prenatal chromosomal microarray testing of fetuses with ultrasound structural anomalies:a prospective cohort study of over 1000 consecutive cases[J]. Journal of Prenatal Diag,2019,39(12):1064-1069.
[12] Bianchi D w, Parker R L, Wentworth J, et al. DNA sequencing versus standard prenatal aneuploidy screening[J].New Engl J Med,2014,371(6):577-578.
[13] Zhang Hailin, Chen Xiaofang, Lv Fangfang, et al. Value of multiplex PCR in the detection of lower respiratory tract infection viruses and atypical pathogens in children [J]. Journal of Wenzhou Medical University.2017.47(11):791-795.

